CRANCOD - A Program for the Analysis and Reconstruction of Spatial Forest Structure
From Arne Pommerening's Forest Biometrics Lab
Contents
System requirements
As with any Java application CRANCOD can be used on Windows, Linux and Macintosh computers:
- Windows XP, Vista, 7/8
- Mac OS X
- Equivalent Linux version
- Java Runtime Environment (JRE)
Installation
- Download the file Crancod.zip onto your computer and unzip it.
- Save the file Crancod.jar on your computer's desktop or in a dedicated target folder.
- If not previously installed download and install the Java Runtime Environment (JRE).
- Launch the CRANCOD program by double clicking on the file Crancod.jar.
CRANCOD Documentation
- Download the user manual here.
- Check out the (slightly) updated poster presentation of CRANCOD at the annual conference of the British Ecological Society in 2006.
CRANCOD Sample input files
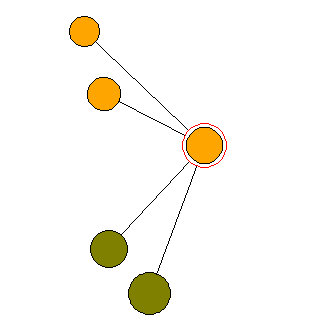
The following data are from my management demonstration plot (MDP) 6 in Clocaenog Forest (North Wales) surveyed in 2004. This is an interesting site where a formerly mono-species Japanese larch plantation has gradually been colonised by Sitka spruce and other species (see the plot map at the top of this page).
- Configuration file Clg6.cfg
- Tree data file Clg6.dat
- Geometry file Clg6.geo
- Species file Clg6.species
CRANCOD Sample output files
These data also refer to MDP 6 in Clocaenog Forest (North Wales) surveyed in 2004.
- Protocol file logging all processes and presenting basic results Clg6_protocol.zip (please unzip)
- File containing all non-spatial results Clg6_NonSpatials_org.out
- File containing all spatial results (NNSS) Clg6_Spatials_org.out
- File with second-order characteristics Clg6_corr_org.out
- File with individual tree results Clg6_tree_org.out
Clg6_protocol.html is an HTML file and can be opened and viewed with any internet browser. All files with the extension *.out are ASCII files and can be opened and viewed in MS Excel.